As Ontario enters a new lockdown, the main concern is a potential overrun of our ICU capacity. A year ago, the Ontario government announced a planned major expansion of provincial ICU capacity, but, given our current limits, it appears that either the plan never came to fruition or that the efforts woefully underestimated possible COVID-19 ICU demand. The Ontario COVID-19 data portal gives the daily ICU COVID-19 census from which we can compute daily changes. As of April 4, Ontario has 451 ICU patients associated with COVID-19. The daily changes give us the net flow but the counting does not tell us how many people actually arrived or departed each day. When we see a daily uptick of 25 people, is that 25 arrivals with no departures or is it 50 arrivals with 25 departures? Both scenarios give the same net change.
Even though the COVID-19 data shows only the net change, we can infer the arrival and departure rates using the Skellam distribution. The Skellam distribution is the difference of two Poisson distributions, ![]() , given by the convolution with a negative sum, namely,
, given by the convolution with a negative sum, namely,
(1) ![]()
where ![]() has support on
has support on ![]() , and
, and ![]() is the Poisson. The resulting distribution can be expressed in terms of the modified Bessel function,
is the Poisson. The resulting distribution can be expressed in terms of the modified Bessel function,
(2) ![]()
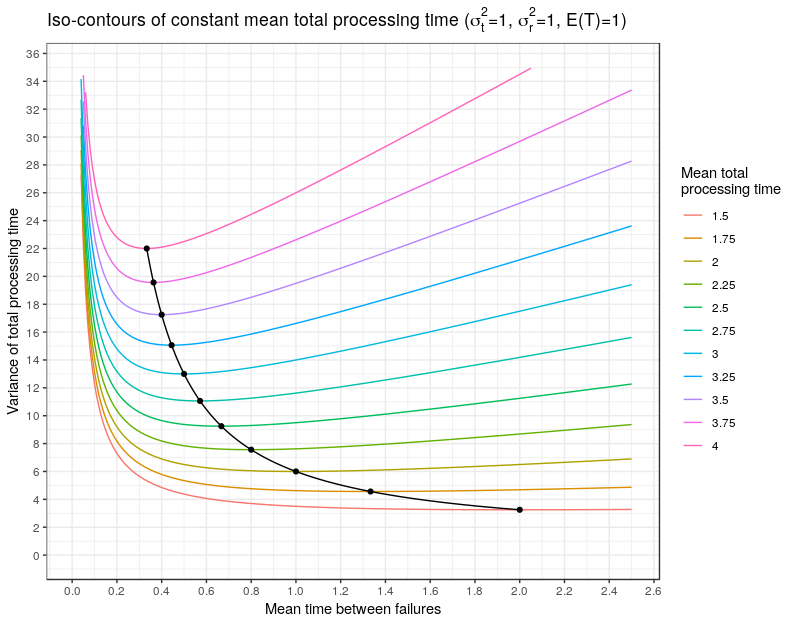
I take the daily difference in the Ontario ICU census data and estimate the Skellam distribution over a 21 day sliding window. The Skellam parameters are the ribbons in figure 1, smoothed with cubic splines with cross validation, and superimposed on the daily net flow. The red ribbon is the arrival process and the blue ribbon is the departure process. Each lollipop point is the net change in the ICU occupation on that day. In figure 2 I show a Q-Q plot of the Pearson residuals from the Skellam model – the model is a reasonably good description of the data generating process. In my analysis I use the R packages Skellam and mgcv (for spline estimation). For clarity, I indicate the lockdown imposition and lockdown lift dates. As of early April 2021, it looks like we are approaching an average of 50 admissions to the ICU every day with an average departure of about 40.

Causation is hard to determine in regressions such as this model, but we see that after the lockdown date on December 26, 2020, the arrival rate quickly plateaued only to begin rising again near the end of the lockdown period. It appears that the lockdown this winter had the effect of tempering arrivals into the ICU. But as always with statistics, we need to be cautious about making causal statements. The arrivals in the ICU are from people who were infected at an earlier date, probably in the neighbourhood of one week earlier. Thus, we see that the arrival rate remains high from the beginning of the lockdown on December 26 through the end of December and early January. We should expect this inertia from infections that occurred near or before the imposition of the lockdown, although there is a distinct possibility that people started to change their behaviour before the lockdown date. But notice that the arrival rate begins to rise sharply almost exactly coincidental with the end of the lockdown on February 16, 2021. This observation suggests that infections were beginning to accelerate at least a week, maybe two, prior to the end of the lockdown. It appears that we would have seen a surge at some level in Ontario’s ICUs even if the lockdown had been maintained through the end of February or longer. Of course it’s quite probable that the resulting surge would have been smaller than the one we are currently witnessing, but to gain insight into such a counterfactual we need to know what caused the acceleration in infections prior to the end of the lockdown. If infections started to rise before the end of the lockdown, is it the result of the new more contagious variants? Is it lockdown fatigue? Is it spread among essential workers? Maybe it’s a combination of effects; I don’t know. Lockdown efficacy rests on that understanding.
Notice that the arrival process turned downward, or at least plateaued, about two weeks after the lockdown date of December 26, 2020, but also notice that the daily number of departures has been steadily increasing throughout the entire winter. Of course we expect that as arrivals increase, departures will eventually increase as well – everyone who enters the ICU eventually leaves. In a steady-state queue, the average departure rate will equal the average arrival rate. With time variation in the arrival process the departure rate will lag as the arrival rate waxes and wanes. It might be instructive to think about the average server load (ICU occupation) of an M/M/c queue (exponentially distributed arrival time, exponentially distributed server times, with c servers) as an approximation to the ICU dynamics. In an M/M/c queue the average number of busy servers (ICU bed occupation) is,
(3) ![]()
where ![]() is the arrival rate and
is the arrival rate and ![]() is the average service rate. For example, given an arrival rate of 50 people per day and an average length of stay of 14 days in the ICU, the steady-state average number of people in the ICU would be 700 patients, provided the number of beds (servers) is larger than 700. I don’t know if the length of stay has been varying over time. It would be great to do a full time dependent competing risk survival analysis on that data, but the steady rise in the departure rate that we see over the winter might indicate that the length of stay in the ICU has also been changing with the arrival rate. I don’t know.
is the average service rate. For example, given an arrival rate of 50 people per day and an average length of stay of 14 days in the ICU, the steady-state average number of people in the ICU would be 700 patients, provided the number of beds (servers) is larger than 700. I don’t know if the length of stay has been varying over time. It would be great to do a full time dependent competing risk survival analysis on that data, but the steady rise in the departure rate that we see over the winter might indicate that the length of stay in the ICU has also been changing with the arrival rate. I don’t know.
There are a number of ways to extend this work. The estimator is crude – instead of a naive 21 day sliding window, it could be built with a Bayesian filter. The Q-Q plot in figure 2 shows that the naive sliding window generates a good fit to the data but if we want to back-out robust estimates of the timing of the infections associated with the arrivals, it might be worth exploring filtering. Second, it is possible to promote the parameters of the Skellam process to functions of other observables such as positive test rates, lagged test rates, time, or any other appropriate regressor and then estimate the model through non-linear minimization. In fact, initially I approached the problem using a polynomial basis over time with regression coefficients, but the sliding window generates a better fit.
Where does this leave us? The precipitous drop in ICU arrivals throughout the summer of 2020 with low social distancing stringency and no lockdowns, along with the steady rise in arrivals beginning in the fall, suggest that Covid-19 will become part of the panoply of seasonal respiratory viruses that appear each year. Hopefully with the vaccines and natural exposure, future seasonal case loads will occur with muted numbers relative to our current experience. Given that the largest mitigating effect, next to vaccines, seems to be the arrival of spring and summer, lockdown or not, the warming weather accompanied by greater opportunities to be outdoors should have a positive effect. Covid-19 cases are plummeting across the southern US states like Texas and Alabama, yet cases are rising in colder northern states like New York and New Jersey. While increased herd immunity in the US from higher levels of vaccination and higher levels of natural exposure must be having an effect on reducing spread, it really does seem like Covid-19 behaves as a seasonal respiratory illness. Hopefully we can expect better days ahead, but, in the end, it is very hard to maintain a large susceptible population in the face of a highly contagious virus that has already become widespread. Perhaps we should have been looking for better public health policies all along.

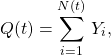
![Rendered by QuickLaTeX.com \begin{align*}\phi_{Q(t)}(u) &=\mathbb{E}[e^{iuQ(t)}] \\ &= \exp\left(rt\ln\left(\frac{1-p}{1-pe^{iu}}\right)\right) \\ &= \left(\frac{1-p}{1-pe^{iu}}\right)^{rt},\end{align*}](https://maybury.ca/the-reformed-physicist/wp-content/ql-cache/quicklatex.com-493e9b3cc164dbdfe835dadda287c3ca_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\mathbb{E}[e^{iuZ(t)}] &= \exp(-t\psi(-\eta(u))) \\ &= \left(1- \frac{r}{b}\ln\left(\frac{1-p}{1-pe^{iu}}\right)\right)^{-at},\end{align*}](https://maybury.ca/the-reformed-physicist/wp-content/ql-cache/quicklatex.com-b8dec93aa84eef4c5279cb4cf8a9011b_l3.png)
![Rendered by QuickLaTeX.com \begin{align*}\mathbb{E}[e^{iuQ(t)}] &= \exp(t\,\eta(u)) \\\mathbb{E}[e^{-sT(t)}] &= \exp(-t\,\psi(s)), \end{align*}](https://maybury.ca/the-reformed-physicist/wp-content/ql-cache/quicklatex.com-a388ec11ca6d71673b206a186378daf0_l3.png)
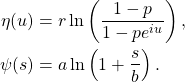
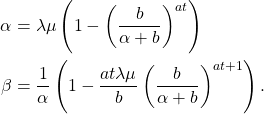



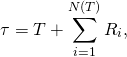
![Rendered by QuickLaTeX.com \begin{align*} \mathbb{E}(\tau|t) &= t + \mathbb{E}\left(\sum_{i=1}^{N(t)} R_i\right) \\ &= t + \sum_{k=0}^\infty \mathbb{E}\left[\left.\sum_{i=1}^k R_i \right| N(t) = k\right]\mathbb{P}(N(t) = k) \\ &= t + \sum_{k=1}^\infty (k\bar r) \frac{(t/\bar f)^k}{k!}e^{-t/\bar f} \\ & = t + \bar r \frac{t}{\bar f}e^{-t/\bar f} \sum_{k=1}^\infty \frac{(t/\bar f)^{k-1}} {(k-1)!} \\ &= t \left(\frac{\bar f + \bar r}{\bar f}\right) \end{align}](https://maybury.ca/the-reformed-physicist/wp-content/ql-cache/quicklatex.com-0916c4d96ee85a712b0d0198d7af0384_l3.png)
![Rendered by QuickLaTeX.com \begin{align*} \mathcal{L}(u) = \mathbb{E}[\exp(uS)] &= \mathbb{E}\left[\exp\left(u\sum_{i=1}^{N(t)} R_i\right)\right] \\ &= \sum_{k=0}^\infty \left[(\mathcal{L}_r(u))^k | N(t) =k \right] \frac{(t/\bar f)^k}{k!}e^{-t/\bar f} \\ &= \exp\left(\frac{t}{\bar f}(\mathcal{L}_r(u) -1)\right), \end{align}](https://maybury.ca/the-reformed-physicist/wp-content/ql-cache/quicklatex.com-578ae091576f08c7ece9087bc7d303b4_l3.png)
![Rendered by QuickLaTeX.com \begin{align*} \mathbb{E}[\text{Var}(\tau|T=t)] &= \mathbb{E}[\text{Var}(t + S)|T=t] \\&= \mathbb{E}[\mathcal{L}^{\prime\prime}(0) - (\mathcal{L}^\prime(0))^2] \\ &= \mathbb{E}\left[ \left(\frac{t\bar r}{\bar f}\right)^2 + \frac{t(\sigma_r^2 + \bar r^2)}{\bar f} - \left(\frac{t\bar r}{\bar f}\right)^2 \right] \\ & = \mathbb{E}\left[\frac{t(\sigma_r^2 + \bar r^2)}{\bar f}\right] = \frac{\bar t(\sigma_r^2 + \bar r^2)}{\bar f} . \end{align}](https://maybury.ca/the-reformed-physicist/wp-content/ql-cache/quicklatex.com-13585279e8c56f48887ace3351029543_l3.png)
![Rendered by QuickLaTeX.com \begin{align*} \text{Var}(\tau) &= \text{Var}(\mathbb{E}(\tau|t) ) + \mathbb{E}[\text{Var}(\tau|t)] \\ & = \left(\frac{\bar r + \bar f}{\bar f}\right)^2\sigma_t^2 + \left(\frac{\bar t}{\bar f}\right) \left(\bar r^2 + \sigma_r^2\right) \end{align}](https://maybury.ca/the-reformed-physicist/wp-content/ql-cache/quicklatex.com-04a53b07232c2f08ab3bb22bf1d49962_l3.png)